Be yourself; Everyone else is already taken.
— Oscar Wilde.
This is the first post on my new blog. I’m just getting this new blog going, so stay tuned for more. Subscribe below to get notified when I post new updates.
Be yourself; Everyone else is already taken.
— Oscar Wilde.
This is the first post on my new blog. I’m just getting this new blog going, so stay tuned for more. Subscribe below to get notified when I post new updates.

Every day, a bevy of information is made available in the field of biology: genetic sequences, patterns in protein activity, medical scans (MRI, PET, CAT, etc.). In order to keep up with the rapid pace of research, and its unparalleled production levels of data, the field of bioinformatics was created.
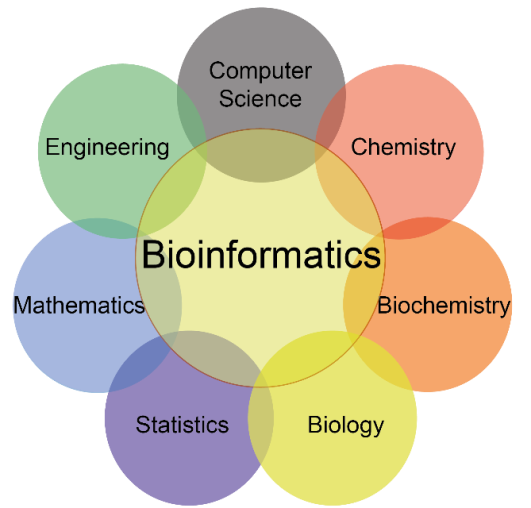
Bioinformatics is an interdisciplinary field of study focused on the acquisition, storage, and archiving of biological data. It centers on the development and application of computational tools to analyze and interpret biological data. These typically comprise mathematical models, computer simulations, and analytical and theoretical methods to study behavioral, molecular, physiological, and social systems.
Below are some common applications of bioinformatics in biomedical research:
As can be seen, one of the most widespread applications of bioinformatics is genomic studies. For this reason, in order to become proficient in this field, it is necessary to have background knowledge in human genetics and the central dogma of molecular biology.
DNA is the primary substance comprising the human genome. It is a complex molecule, in the form of a double helix, whose individual units are known as nucleotides. Each nucleotide is composed of a Nitrogen base (Adenine, Thymine, Guanine, and Cytosine), a sugar molecule (deoxyribose), and a Phosphate group. The two strands are linked together by hydrogen bonds between the bases (Adenine with Thymine and Guanine with Cytosine). The proportion of Cytosine-Guanine pairs found in a sample determines the stability of the sample (if there is more Cytosine-Guanine), then the sample is said to be more stable.
The human genome has about 3.1 billion base pairs, amounting to about 10 Terabytes of data. In an era where big data has become a buzzword and has skyrocketed in importance, the genome is a definite object of the computational advances currently being made.
The central dogma of molecular biology, which is the core mechanism behind genomic studies and data, consists of the process in which DNA is used to manufacture protein.
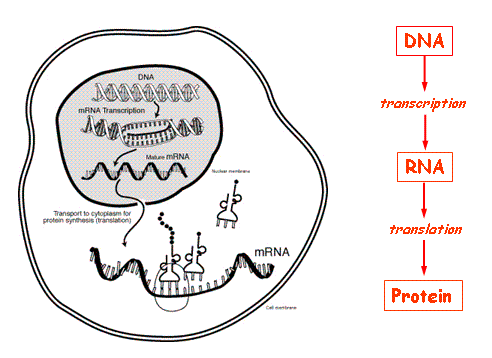
As can be seen above, this process begins with transcription, in which mRNA (messenger RNA, which is a nucleic acid serving as an intermediary between DNA and protein) is produced from the template strand. This sequence is later translated into polypeptide sequences in the ribosomes, which aggregate and fold into proteins, which, in turn, carry out the functions of the cell. Polypeptides are composed of chains of amino acids, which are coded for by individual units of RNA (and DNA before RNA) called codons ( units of 3 nucleotides). A single amino acid can be coded for by multiple codons, in a phenomenon known as the wobble effect. Thus, when a mutation (general term used to describe a change in genetic information) occurs in the DNA or RNA, it may not have any effect on the resulting protein.

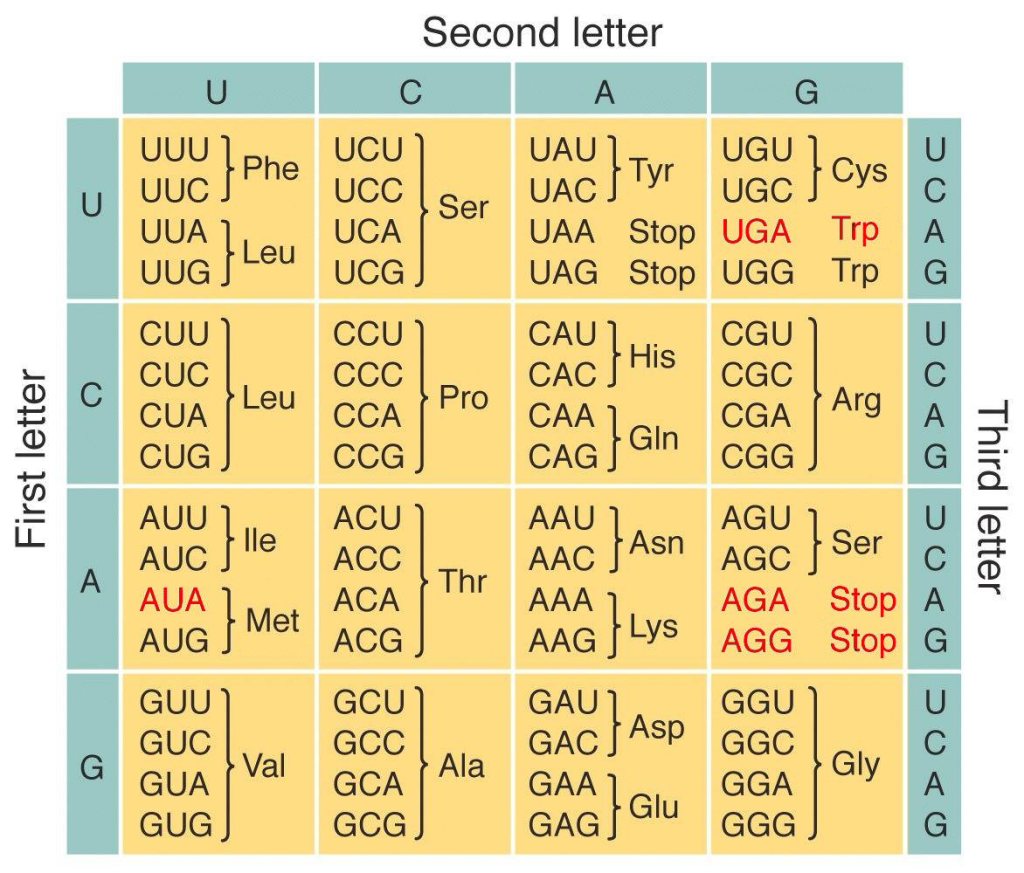
Interestingly, the corresponding amino acid for each codon seems to be very similar in every living organism, the reason for which it is often referred to as the universal genetic code.
This mechanism encodes every life process. The purpose of bioinformatics is to improve the understanding of these processes, starting at their beginning point, in order to better exploit them or understand the mechanisms behind their degradation (i.e. as in cancer).
This is an example post, originally published as part of Blogging University. Enroll in one of our ten programs, and start your blog right.
You’re going to publish a post today. Don’t worry about how your blog looks. Don’t worry if you haven’t given it a name yet, or you’re feeling overwhelmed. Just click the “New Post” button, and tell us why you’re here.
Why do this?
The post can be short or long, a personal intro to your life or a bloggy mission statement, a manifesto for the future or a simple outline of your the types of things you hope to publish.
To help you get started, here are a few questions:
You’re not locked into any of this; one of the wonderful things about blogs is how they constantly evolve as we learn, grow, and interact with one another — but it’s good to know where and why you started, and articulating your goals may just give you a few other post ideas.
Can’t think how to get started? Just write the first thing that pops into your head. Anne Lamott, author of a book on writing we love, says that you need to give yourself permission to write a “crappy first draft”. Anne makes a great point — just start writing, and worry about editing it later.
When you’re ready to publish, give your post three to five tags that describe your blog’s focus — writing, photography, fiction, parenting, food, cars, movies, sports, whatever. These tags will help others who care about your topics find you in the Reader. Make sure one of the tags is “zerotohero,” so other new bloggers can find you, too.